TOOLS REQUIRED
Tools of Recombinant DNA Technology
- The key tools required for the recombinant DNA technology are:
(i) Restriction enzymes(ii) Polymerase enzymes
(iii) Ligases(iv) Vectors
(v) Host organism/cell.
Restriction Enzymes
- The restriction enzymes are called “molecular scissors” and are responsible for cutting DNA.
- They are present in bacteria to provide a type of defence mechanism called the “restriction-modification system”.
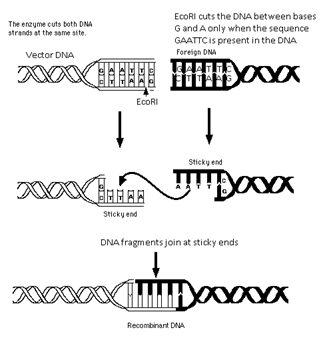
- The first restriction endonuclease, HindII, was isolated by Smith, Wilcox and Kelley (1968) from Haemophilus influenza bacterium. It was used to cut DNA molecules at a particular point by recognizing a specific sequence of six base pairs, known as the recognition sequence.
Naming of Restriction Enzymes:
- Restriction enzymes belong to a class of enzymes called nucleases and are of two types:
(i) Exonucleases – cut DNA at the ends.
(ii) Endonucleases – cut at specific positions within the DNA.
- The recognition sequences of endonucleases are palindromic, i.e., the sequence of base pairs read the same on both the DNA strands, when orientation of reading is kept same, e.g.,
5’ ______ GAATTC ______ 3’
3’ ______ CTTAAG ______ 5’
Mechanism of Action of Endonucleases
- Every endonuclease inspects the entire DNA sequence for the palindromic recognition sequence.
- On finding the palindrome, the endonuclease binds to the DNA.
- It cuts the opposite strands of DNA in the sugar – phosphate backbone; a little away from the centre of the palindrome sites but between the same bases on both strands.
- This results in the formation of single stranded overhanging stretches at the end of each strand called sticky ends.
- The sticky ends facilitate the action of the enzyme DNA ligase by readily forming hydrogen bonds with complementary strands.
- In genetic engineering, DNA fragments have same kind of sticky ends.
- These sticky ends are complementary to each other and thus can be joined by DNA ligase (end – to – end).
Related Keywords
 SureDen
SureDen